Effect of climate change on food web processes
Effects of climate change on the food-web structure can have large consequences for the bioaccumulation of POPs. These can be bottom-up effects, in which changes in the abundance of primary producers influence the relative abundance of biota at higher trophic levels. They can also be top-down effects, where changes in the abundance of predators influence the relative abundance of organisms at lower trophic levels. Some top-down effects may be more evident, as changes in ice cover will have devastating effects on the abundance of top predators that depend on ice, such as polar bears and seals. Other top-down effects, such as predators switching to different prey and species moving to new regions, are more difficult to predict. Climate change effects on processes related to the intake of POPs could be enhanced intake in gills and intestines due to warmer temperatures and increased metabolism which may decrease internal concentrations or lead to increased exposure to toxic metabolites. Enforced fasting for e.g. polar bear may lead to increased residue levels.
Impacts on bioaccumulation modelling
There could be three primary mechanisms through which climate change can affect the bioaccumulation of environmental contaminants (Gouin et al., 2013):
- climate change induced changes in environmental exposure (M1)
- dietary exposure (M2) and
- species-to-species uptake and loss rates (M3)
To date, only few modelling studies have investigated these mechanisms to some extent in the Arctic and Baltic Sea regions. In general, the physical environment played a relatively small role in determining the susceptibility to chemical exposure compared to the food web properties.
Hence, the global climate change induced direct changes (temperature and precipitation) would have little influence on the bioaccumulation of POPs in foods consumed by humans. Climate change induced effects were most likely to be indirect, such as changes in the dietary composition of food chains (M2). The direct effects on temperature-dependent uptake and loss rates are not likely to be significant compared to the indirect effects (M3).
Marine Food Web Modelling
A marine food web was chosen from the Svalbard region for initial model development. The Svalbard food web model contains plankton and fish samples taken within the ArcRisk project. A review of the marine ecosystem of Kongsfjorden in Svalbard by Hop et al. (2002) was utilized to help shape the initial structure of the Svalbard marine food web. The species included in the food web were algae, copepods (Calanus hyperboreus), amphipods (Onisimus spp., Apherusa glacialis and Themisto libellula), polar cod (Boreogadus saida), three seal species (Erignathus barbatus, Phoca hispida and P. groenlandica), and polar bears (Ursus maritimus). Levels of PCB in Svalbard waters in 2001 were used in the model (Sobek and Gustafsson, 2004).
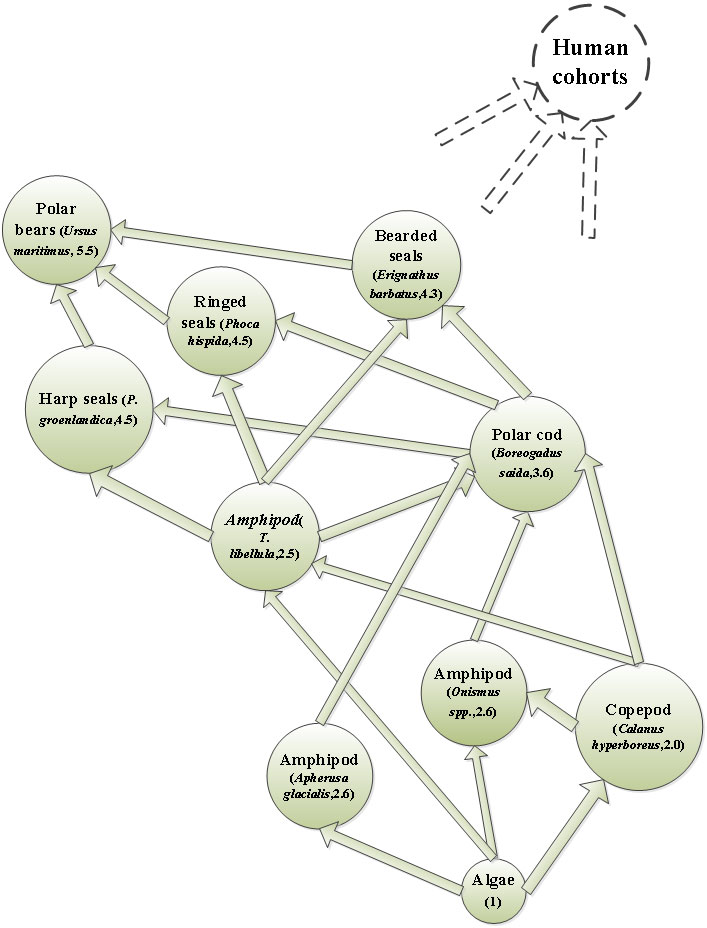
The food web model underestimates the measured concentrations, which vary with the species and the PCB congener. PCB concentrations predicted in polar cod were most sensitive to the digestion factor, which suggests biomagnification as a very important factor. The herbivorous Apherusa glacialis are more affected by PCB concentrations in the water.
Humans living on Svalbard are relatively few in number (~2500), have a short residence time there and their diet does not include large contribution from local marine fish or mammals. Hence, studies of human exposure were done in other Arctic areas instead. The food web model from Svalbard is still highly relevant for human exposure in other Arctic areas, e.g. since the surrounding Barents Sea is important for the Norwegian and Russian fishing industry and the consumers. Bioaccumulation key processes are similar around the Arctic. Hence, results and tools developed within the Svalbard food web model can be applied on other Arctic food web systems that are more relevant for human exposure. However, even while using state-of-the-art modelling tools, the estimations of bioaccumulation in Arctic marine food webs under a changing climate is very difficult. Potential changes in the ecosystem, impact of invading species and ocean acidification are yet not fully understood.
Due to the slow uptake and elimination rates of many POPs at higher trophic levels (marine mammals), a dynamic model could provide improved predictions compared to a static model. A dynamic model is also expected to be more suitable compared to a steady-state model regarding PFAS, since the PFAS exposure changes rapidly over time.
To develop a relevant future climate change scenarios for food web modelling is challenging. A review by Cousins et al. (2011) demonstrated that predicting changes in food web structure due to future climate change is not yet possible. Information about possible scenarios and already on-going changes observed are needed before models can be further developed for food webs in change. ArcRisk have developed bioaccumulation models for Arctic food webs. However, data of POPs in sea water is still scarce, although needed to improve the models.





















